
[ad_1]
CRISPR-Cas9 is a bacterial adaptive immune response system that has been harnessed as a exact genome enhancing instrument. CRISPRs had been first recognized in E. coli in 1987, however the position of CRISPRs bacterial immunity was not recognized till the early 2000s. In 2012, a landmark paper displaying CRISPR-Cas9 as a gene-editing instrument was revealed. In 2020, The Nobel Prize in Chemistry was awarded to Emmanuelle Charpentier and Jennifer Doudna for his or her improvement of this highly effective gene-editing instrument.
On this article, we share how CRISPR was first recognized, the important thing discoveries that led to its improvement as a gene-editing instrument, uncover the astounding makes use of of this instrument, and see the way it has continued to be developed past gene-editing.
CRISPR-Cas9 within the Highlight
CRISPR-Cas techniques, equivalent to CRISPR-Cas9, are adaptive immune response techniques that defend prokaryotes from bacteriophages. They work by cleaving the nucleic acids of invading viruses, thus defending prokaryotes from viral infections.
The usage of CRISPR-Cas9 to edit genes was thrust into the highlight in 2012 when George Church, Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang harnessed it as a instrument to change focused areas of genomes. Given its potential to revolutionize gene enhancing, Science named CRISPR Breakthrough of the Yr in 2015. In 2020, this expertise earned Jennifer Doudna and Emmanuelle Charpentier the Nobel Prize in Chemistry.
However how was CRISPR first recognized? When was its position in bacterial immunity found, and what different steps led to the event of this instrument? Determine 1 reveals an outline of among the key occasions within the historical past of CRISPR, and we focus on these in additional element within the following sections.
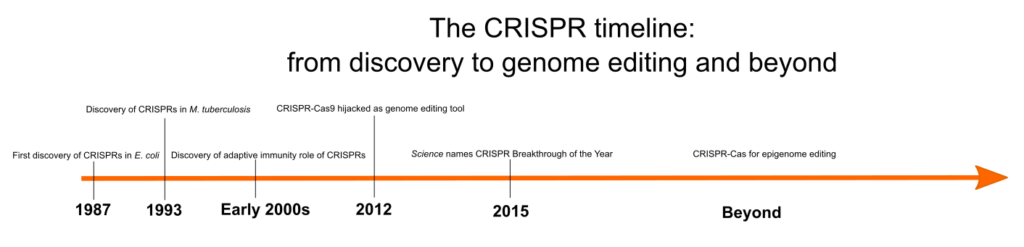
Discovery of CRISPRs
Clustered regularly interspaced short palindromic repeats, in any other case often called CRISPRs, are repeating DNA sequences within the genomes of prokaryotes, equivalent to micro organism and archaea. CRISPRs had been first recognized in E. coli in 1987 by a Japanese scientist, Yoshizumi Ishino, and his group, who by chance cloned an uncommon sequence of repeated sequences interspersed with spacer sequences whereas analyzing a gene chargeable for the conversion of alkaline phosphatase. [1] Nonetheless, as a result of lack of adequate DNA sequence knowledge, the perform of those arrays remained a thriller.
In 1993, researchers led by J.D. van Embden within the Netherlands found that completely different strains of Mycobacterium tuberculosis had completely different spacer sequences between the DNA repeats. They characterised M. tuberculosis strains based mostly on their spacer sequences, a way often called ‘spacer oligonucleotide typing’ or spoligotyping. [2] Subsequently, these sequences had been recognized in a number of different bacterial and archaeal genomes. Researchers Francisco Mojica and Ruud Jansen had been the primary to consult with them as CRISPRs. [3]
CRISPR-Cas Programs as an Adaptive Immune Response
When CRISPR techniques had been first found, they had been regarded as a novel DNA restore mechanism in thermophilic archaea and micro organism. [4] Within the early 2000s, Mojica and coworkers observed that the spacer sequences had been much like sequences present in bacteriophages, viruses, and plasmids. They found that viruses can not infect micro organism possessing homologous spacer sequences, suggesting that these sequences play a task within the adaptive immune system in prokaryotes. [5]
In 2007, Rodolphe Barrangou, together with different researchers at Danisco, revealed a paper displaying that new spacers derived from genomic sequences of phages had been built-in into the virus-challenged micro organism. Additionally they demonstrated that eradicating or including specific spacers altered the phage resistance of the cell. [6]
How the CRISPR Adaptive Immune Response Works
When a virus infects a prokaryote, the spacer sequences in CRISPR arrays are transcribed to generate brief CRISPR RNA (crRNA), which guides the CRISPR-associated sequence (Cas) protein to cleave complementary DNA or RNA viral sequences, relying on the kind of CRISPR-Cas system. On this approach, CRISPR-Cas techniques perform as a protection mechanism to stop repeated infections by the identical virus.
Different Elements of CRISPR-Cas Programs
The position of Cas proteins as nucleases that cleave at particular websites was found by Makarova and colleagues, who performed a comparative genomic evaluation of CRISPR and Cas genes. They predicted that the perform of CRISPR-Cas techniques was much like that of RNA interference, through which protein complexes silence genes by cleaving mRNA. [6] Whereas some Cas proteins cleave DNA, others cleave RNA. For instance, Cas9 cleaves DNA, whereas Cas13 cleaves RNA.
Along with Cas proteins, protospacer adjacent motifs (PAMs) are important parts of CRISPR-Cas techniques. Because the title suggests, PAMs are brief 2–6 base-pair sequences within the viral genome discovered adjoining to sequences focused by Cas nucleases. Cas nucleases acknowledge PAMs and can’t cleave DNA except a PAM is current. PAMs play an vital position in making certain that solely overseas viral nucleic acids are cleaved however not CRISPR arrays. [8]
CRISPR-Cas9 Hijacked for Genome Enhancing
In 2012, George Church, Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang found that by designing information RNA to focus on a selected area within the genome, “the CRISPR-Cas9 system can be utilized as a “cut-and-paste” instrument to change genomes. As a DNA-editing instrument, CRISPR-Cas9 can take away or introduce new genes in addition to silence or activate genes. CRISPR-Cas9 has been used to change off genes that restrict the manufacturing of lipids in microalgae, resulting in elevated lipid manufacturing and better yields of biofuel. [9] Within the close to future, CRISPR-Cas9 can also be used to treatment genetic problems equivalent to sickle-cell anemia and cystic fibrosis. [10] In actual fact, there’s already a variety of CRISPR functions in illness therapy, together with most cancers and infectious ailments.
A New Wave of CRISPR Instruments
Since CRISPR’s debut as a genome enhancing instrument, it has been tailored in varied artistic and useful methods. Level mutations can be utilized to inactivate one of many catalytic domains in Cas9, producing nickases, which reduce only one strand of double-stranded DNA. Pairing two completely different nickases collectively can obtain the identical double-strand break as absolutely energetic Cas9 however with enhanced specificity. [11]
Researchers have modified the Cas9 nuclease to carry out focused epigenome enhancing. This modified Cas9, often called enzymatically useless Cas9 (dCas9), could be linked to certainly one of a number of enzymes that change the epigenome, e.g., DNA demethylases, methylases, or acetyltransferases. Like unmodified Cas9, dCas9 is directed to the focused genomic area with a information RNA. Nonetheless, as a substitute of slicing the DNA, the dCas9-enzyme complicated modifies the epigenome on the web site.
Epigenome enhancing can activate or repress transcription. Transcription could be activated by demethylating DNA utilizing enzymes equivalent to dCas9-Tet1 or by modifying histones utilizing dCas9 linked to the histone acetyltransferase p300 enzyme. Conversely, transcription could be repressed by methylating DNA utilizing DNA methyltransferase. Genes can be silenced by linking dCas9 to enzymes that recruit corepressor proteins.
Equally, gene expression could be activated (CRISPRa) or inhibited (CRISPRi) by fusing dCas9 to transcriptional effectors that activate or repress transcription.
Cas nucleases have additionally been engineered to lower the extent of off-target exercise, producing high-fidelity Cas9 variants equivalent to SpCas9-HF1 and eSpCas9-1.1.
Different CRISPR Nucleases
Cas9 remoted from Streptococcus pyogenes is the best-known and most well-used nuclease, however different nucleases have been recognized which have completely different properties. Cas9 nucleases from different species have completely different PAM specificities and sizes, which may impression the benefit of use.
Different flavors of Cas nucleases have additionally been discovered, together with Cas13, which can be utilized to trace, modify, and knockdown RNA in cells, and Cas12, which is extra compact and higher for enhancing AT-rich websites than conventional Cas9.
CRISPR within the Clinic
One of many apparent potentials of CRISPR is within the therapy of genetic ailments. This potential has been reached with the transfer of CRISPR from the analysis bench to the clinic as a therapeutic instrument.
In 2019, CRISPR enhancing was trialed as a therapy for sufferers affected by sickle cell illness and β-Thalassemia. [12] This success has led to many extra sufferers receiving the identical therapy.
And in 2023, CRISPR was used to change CAR-T cells to deal with youngsters with relapsed T-cell acute lymphoblastic leukemia. [13]
The Way forward for CRISPR-Cas9
CRISPR-Cas9 expertise has its professionals and cons. Nonetheless, as researchers proceed to make use of, refine, and increase the CRISPR toolkit, new functions and extra instruments are continually arising.
Whether or not you need to know methods to get began with CRISPR or discover new functions of this genome enhancing instrument, head over to the Bitesize Bio CRISPR Analysis Hub.
CRISPR has revolutionized gene enhancing. And there’s loads to be taught. Get your free Gene Enhancing 101 eBook to stand up to hurry immediately.
Initially revealed June 2020. Reviewed and up to date November 2023.
References
- Ishino, Y. et al. Historical past of CRISPR-Cas from encounter with a mysterious repeated sequence to genome enhancing expertise. Journal of Bacteriology, 200, 7 (2018). e00580-17. doi: 10.1128/JB.00580-17.
- Sola, C. et al. Excessive-throughput CRISPR typing of Mycobacterium tuberculosis complicated and Salmonella enterica serotype Typhimurium. Strategies in Molecular Biology, 1311 (2015). doi:10.1007/978-1-4939-2687-9_6.
- Morange, M. et al. What historical past tells us XXXVII. CRISPR-Cas: The invention of an immune system in prokaryotes. Journal of Biosciences, 40 (2015). 221–223 https://doi.org/10.1007/s12038-015-9532-6.
- Makarova, KS. et al. A DNA restore system particular for thermophilic Archaea and micro organism predicted by genomic context evaluation. Nucleic Acids Analysis, 30, 2 (2002). 482–496. doi: 10.1093/nar/30.2.482.
- Mojica FJ, et al. Intervening sequences of frequently spaced prokaryotic repeats derive from overseas genetic parts. J Mol Evol. 2005 Feb;60(2):174-82. doi: 10.1007/s00239-004-0046-3.
- Barrangou R, et al. CRISPR supplies acquired resistance towards viruses in prokaryotes. Science. 2007 Mar 23;315(5819):1709-12. doi: 10.1126/science.1138140.
- Makarova, KS. et al. A putative RNA-interference-based immune system in prokaryotes: computational evaluation of the anticipated enzymatic equipment, practical analogies with eukaryotic RNAi, and hypothetical mechanisms of motion. Biology Direct, 1, 7 (2006). doi: 10.1186/1745-6150-1-7.
- Shah, SA. et al. Protospacer recognition motifs: combined identities and practical variety. RNA Biology, 10, 5 (2013), 891–899. doi: 10.4161/rna.23764.
- Sharma, P.Okay. et al. Tailoring microalgae for environment friendly biofuel manufacturing. Frontiers in Marine Science, 5 (2018), 382. doi: 10.3389/fmars.2018.00382.
- Kotagama, O.W. et al. Period of genomic medication: a story evaluation on CRISPR expertise as a possible therapeutic instrument for human ailments. Biomed Analysis Worldwide, 2019 (2019). doi: 10.1155/2019/1369682.
- Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome enhancing specificity. Cell. 2013 Sep 12;154(6):1380-9. doi: 10.1016/j.cell.2013.08.021.
- Frangoul H, et al. CRISPR-Cas9 Gene Enhancing for Sickle Cell Illness and β-Thalassemia. N Engl J Med. 2021 Jan 21;384(3):252-260. doi: 10.1056/NEJMoa2031054. Epub 2020 Dec 5. PMID: 33283989.
- O’Leary Okay. Base-edited CAR T cells for pediatric leukemia. Nature Drugs. 2023 Jun 22. doi: 10.1038/d41591-023-00056-0.
[ad_2]