
[ad_1]
Genetic modifying is essential to know human illness. We often use immortalized cells for in vitro experiments, however they’ve irregular genomes that don’t characterize the typical human. Induced pluripotent stem cells will be differentiated into nearly any sort and genomically edited with CRISPR, creating a strong different to immortalized cell strains to check human biology.
Tailored from Exact, Excessive-Effectivity Modifying of Stem Cells to Probe Human Biology and Mannequin Illness, a BitesizeBio Webinar with Invoice Skarnes.
Learning human illnesses and genetic variation isn’t straightforward. In vitro research permit us to control the genome of cells fairly simply, however creating mannequin programs which are straightforward to work with and provides legitimate information is difficult.
To beat this, we often resort to immortalized cells. However these cell strains should not regular. They’ve vital mutations on their genome, exhibit attributes not present in wholesome in vivo cells, and a few have unethical origins.
Luckily, there’s a substitute for immortalized cells.
Utilizing CRISPR to edit induced pluripotent stem cells is a approach to create physiologically related and translational fashions. That’s what we’ll talk about on this article.
We’ll make the fundamentals extraordinarily clear and clarify the guiding rules of genome modifying matters. Plus, we offers you a heap of sensible steps to observe and enhance modifying effectivity, design information RNAs, management the zygosity of your clones, and extra!
The CRISPR iPSC Genome Modifying Platform to Probe Human Biology
This sensible method to exploring human biology and modeling illness combines two applied sciences which have gained Nobel Prizes. These are:
- Cell reprogramming.
- CRISPR.
We’ll delve just a little deeper into these subsequent. However briefly, cell reprogramming allows the creation of induced pluripotent stem cells (iPSCs) from somatic cells, and, utilizing CRISPR, we are able to edit the genome of iPSCs to vary their phenotype.
This offers us entry to an unlimited quantity of human biology. For instance:
- Early human growth (embryoids).
- Organogenesis (organoids).
- Hematopoiesis/innate immunity.
- Uncommon illnesses.
- Gene remedy.
- Cell biology.
- Metabolomics.
Now let’s study these applied sciences in just a little extra element.
Induced Pluripotent Stem Cells
The ability of iPSCs is that they are often differentiated into nearly any cell sort of the human physique.
Plus, these cells have regular genomes, in contrast to cells generally used prior to now to handle human cell biology and biochemistry, equivalent to HeLa and HEK293 cells, which have extremely irregular genomes that don’t characterize that of the typical particular person.
And genomic abnormality apart, the non-consensual origin of HeLa cells raises a critical moral situation.
Moreover, you may tradition iPSCs indefinitely and preserve a traditional diploid karyotype: a wholesome set of 23 homologous pairs of chromosomes.
There are some disadvantages to iPSCs. The primary ones are that the effectivity of inducing pluripotency is usually low and entails the viral supply of typically carcinogenic genes into somatic cells.
For a better take a look at the benefits and disadvantages of iPSCs and the way they’re ready, try this paper by Medvedev, ShevchenkI, and Zakian. [1]
What Is CRISPR?
CRISPR is a revolutionary gene modifying expertise that wants little introduction to most readers. If you happen to want an introduction, try Chunkmeasurement Bio’s straightforward information to CRISPR.
In brief, it’s a strong instrument to chop and alter DNA sequences each in vitro and in vivo. A small RNA molecule referred to as information RNA (gRNA) guides a nuclease (often Cas9) to a desired goal location within the genome to take away or add genes, appropriate mutations, and even regulate gene expression.
CRISPR presents two salient benefits over typical gene modifying instruments. These are:
- You can also make single base pair edits by chopping or nicking DNA. [2]
- You possibly can modify each copies of a gene concurrently (biallelic mutations).
Nonetheless, for CRISPR to be a legitimate genome modifying instrument, there are two important necessities:
- The goal sequence meant for modification is genomically distinctive.
- The goal sequence is a number of base pairs upstream of a protospacer adjoining motif sequence (a PAM sequence).
If both of those standards just isn’t met, CRISPR is not going to work.
Try this put up to be taught extra about Cas9s and what strands they nick.
What Are PAM Sequences?
PAM sequences are brief sequences of DNA which are important for cleavage by a Cas nuclease.
They’re situated on the non-complementary strand: the nucleic acid strand, with the similar sequence because the information RNA.
And they’re usually 3–4 base pairs downstream of the Cas nuclease lower website.
You’ll discover a listing of widespread PAM sequences in Bitesize Bio’s information to nucleases and in the direction of the underside of this Addgene article on CRISPR.
A phrase to the clever—when designing guides to your CRISPR experiments, keep away from ones that introduce mutations into the PAM sequence.
Sensible Steerage for CRISPR iPSCs Genome Modifying Tasks
Now you recognize the advantages of making a substitute for immortalized cells utilizing iPSCs and CRISPR, listed here are some sensible tips about the best way to do it nicely.
Guiding Rules
5 primary rules or steps that apply to any genome modifying venture.
1. Subclone the iPSCs
Subcloning induced pluripotent stem cells (iPSCs) allows you to isolate a clonal inhabitants of cells with the most effective genetic properties to your venture.
Doing so will scale back the genetic heterogeneity of your edited clones, rising the consistency and reproducibility of your outcomes.
Moreover, subcloning helps remove undesirable off-target results that will come up from the genome modifying course of.
2. Cautious Characterization of the iPSCs
It’s at all times clever to characterize your iPSC line to make sure they’ve:
- An intact genome. Use karyotyping, high-density SNP arrays (mentioned later), and whole-genome sequencing.
- Unmutated TP53. It usually turns into mutated in cultured iPSCs.
- Good differentiation potential. Use an mRNA microarray, RNA sequencing, or qPCR. [3]
3. Edit the Genome
Introduce mutations that relate solely to your research: ones discovered within the human inhabitants with no further adjustments.
Though you may introduce silent mutations which enhance modifying effectivity, these may influence gene transcription and trigger undesirable off-target results.
4. Characterize the Submit-Edit Clones
Characterize your post-edit clones to verify if in case you have launched your required mutation.
Additionally, verify the cells to make sure they keep a traditional genome after modifying. Search for off-target results you will have by accident launched as a consequence of your required mutation.
Karyotype the cells once more and use long-range PCR to examine high-density SNP arrays to find out the zygosity of your clones (extra on this later).
5. Management/Reversion
When you establish the cell phenotype you want to show is because of the mutation, not one thing else acquired throughout culturing and modifying, revert it to the wild-type sequence utilizing CRISPR (Determine 1).
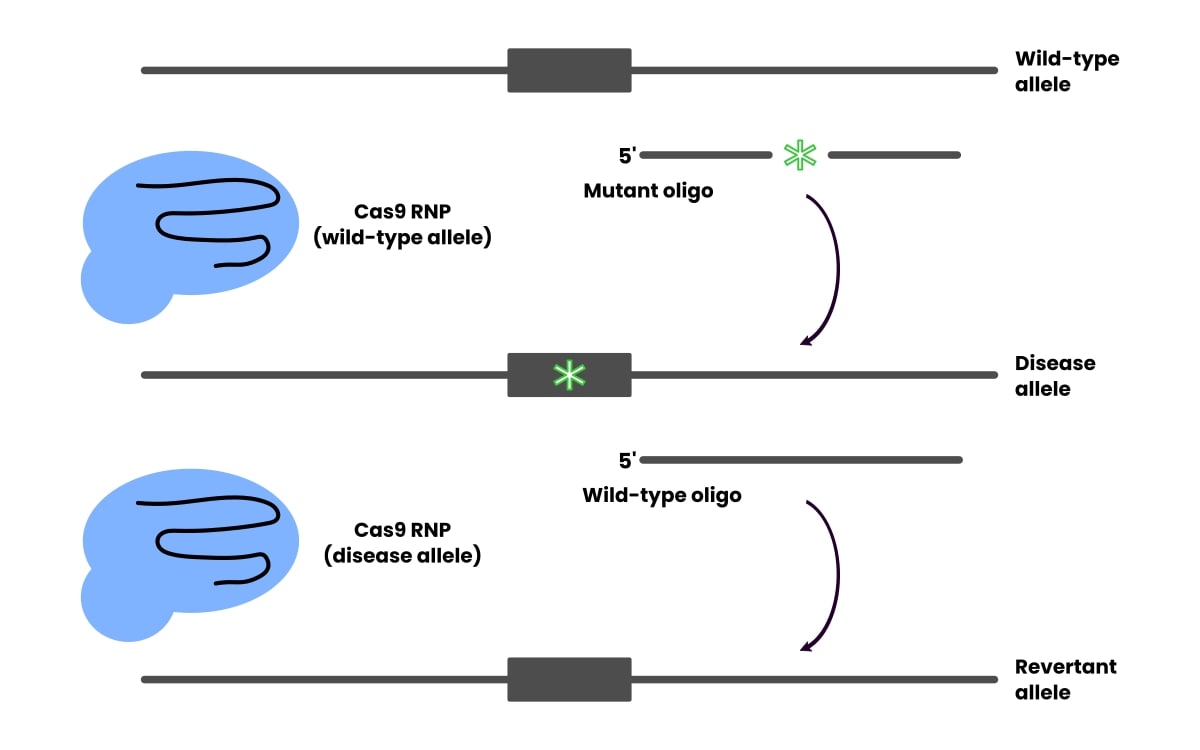
Reversion controls for any off-target results, however remember that the revision effectivity doesn’t essentially correlate to the modifying effectivity.
DNA Restore Strategies in CRISPR
RNA serves as a information to direct the Cas nuclease to particular goal DNA sequences, and the RNA–nuclease complicated known as a ribonucleoprotein (RNP).
These complexes acknowledge a particular facet of the genome and make a double-stranded break.
You should use both of two pathways to restore the break:
- Non-homologous finish becoming a member of.
- Homology-directed restore.
Non-homologous finish becoming a member of (NHEJ) is an error-prone course of that ends in insertion/deletion mutations on the break website on one or each alleles. That is helpful for initiatives the place you need to mutate the gene in such a approach.
Homology-directed restore (HDR) entails including a donor template that can get integrated on the breakpoint and introduce single nucleotide variants.
Try Determine 2 to see this represented graphically.
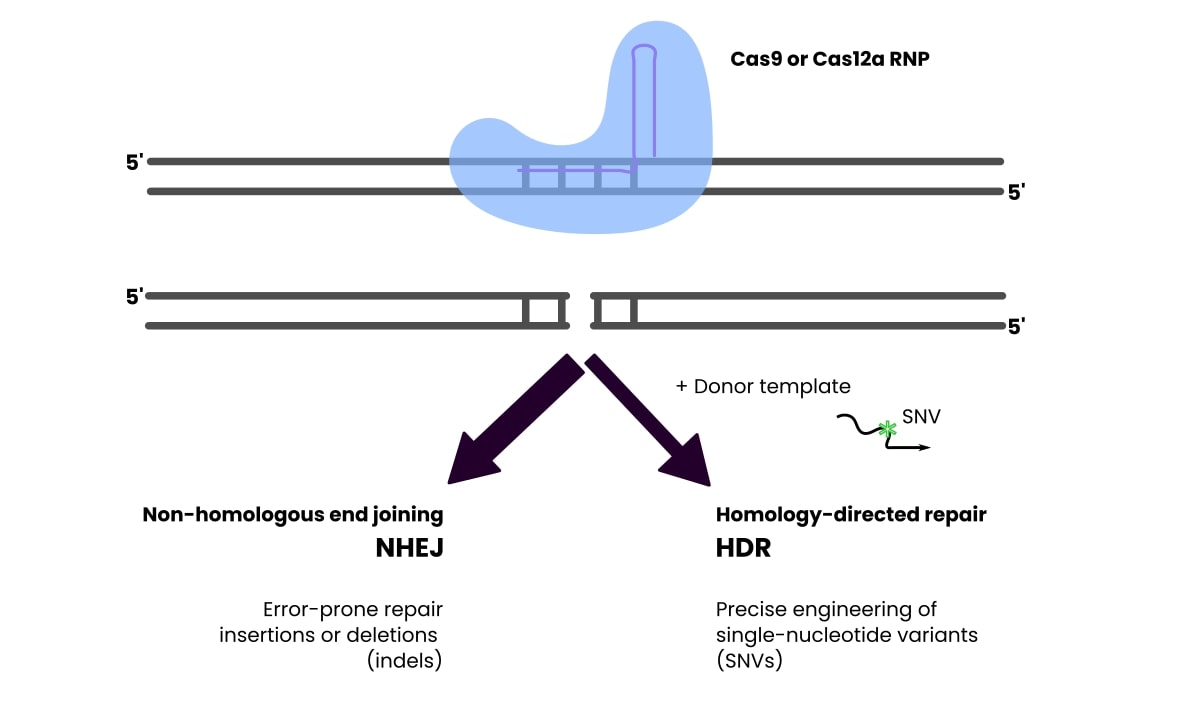
The 2 strategies compete with one another. HDR is exact however will be inefficient, with a low share of the cells exhibiting the specified mutation. This may very well be an issue for large-scale initiatives.
NHEJ is extra environment friendly however liable to the imprecise mutation sorts talked about above.
Methods to Enhance the Effectivity of HDR
As talked about, the nuclear an infection effectivity for HDR will be low. Maybe decrease than 10%. Luckily, there are methods to enhance its effectivity. Methods to extend its effectivity embody:
- Small molecule enhancers. [4]
- Chilly shock. [5]
- Finish-modified oligo donors. [6]
The good thing about these strategies is that they contain delivering reagents and situations to the cells. No mobile choice or enrichment is required.
And observe that this listing isn’t exhaustive. You possibly can try an outline of methods right here. [7]
Easy methods to Monitor the Effectivity-Bettering Methods
So that you’ve acquired some concepts on enhancing your gene modifying effectivity, however how do you monitor them to know if they’ve labored?
If you happen to change the C to a T within the histidine codon of blue fluorescent protein BFP, you get inexperienced fluorescent protein GFP.
- CAT → Histidine.
- TAT → Tyrosine.
These proteins have completely different spectral properties; the previous is blue, the latter inexperienced.
You possibly can introduce this mutation utilizing CRISPR underneath completely different iPSCs situations to observe the effectivity of the modifying course of and choose situations that maximize modifying effectivity.
This can be a easy but highly effective assay to observe the effectivity of NHEJ and HDR. Plus, the outcomes are straightforward to interpret! [8,9]
- No mutation → blue fluorescence.
- Profitable HDR mutation → inexperienced fluorescence.
- Profitable NHEJ mutation → no fluorescence.
The explanation that NHEJ will lead to no fluorescence is due to the insertion or deletion mutations. It may even lead to frameshifts when the size of the insertion or deletion just isn’t a a number of of three.
Designing Information RNAs
When doing genome modifying, you need to design information RNAs that get the Cas nuclease to the specified goal website on the genome.
WGE is a wonderful CRISPR information RNA design instrument that helps establish guides that overlap the goal website and reveals you their place relative to the PAM sequence. [10]
It additionally provides an off-target effectivity rating that quantifies the variety of off-target websites with X base variations.
Purpose for guides that haven’t any matches to different areas on the genome and only a few one- or two-base mismatches.
And, as talked about, keep away from guides that mutate the PAM sequence.
Display Information RNAs in vitro and in vivo
Guides are the wild playing cards in genome modifying experiments, and there can be variable exercise between guides.
A information could have a wonderful off-target rating however solely return wild-type clones or work for less than (say) NHEJ. An nearly an identical information (say simply offset just a little bit to the primary one) could return ample HDR base-edited cells with the specified mutation.
Generally, a number of or extra potential information RNAs will information the Cas9 nuclease to the goal sequence for cleavage.
When that is the case, display them in vitro utilizing a mobility shift assay to see which guides cleave the goal sequence greatest.
Then, whenever you come to make use of guides for genome modifying experiments, tabulate your sequencing outcomes to see which guides provide you with homozygous biallelic mutations, heterozygous mutations, and no mutations (wild-type).
You might also want to take a look at if the guides work for HDE, NHEJ, or each.
You might uncover a information will solely work for one restore technique. This offers you an perception into how nicely a given mutation is tolerated or whether it is deadly. For instance, no mutations had been launched by way of HDR, however you get considerable wild-type and profitable NHEJ-edited clones.
Easy methods to Management the Zygosity of Clones
Relying on what you’re learning (particularly if you’re engaged on genetic illnesses), it’s possible you’ll want to management the zygosity of your edited clones.
In different phrases, management whether or not you introduce the mutation to at least one or each alleles.
An efficient technique is perhaps so as to add inactivated Cas9 (additionally referred to as lifeless Cas9 or dCas9) with the information RNA throughout the nuclear an infection step. [9]
The dCas9 will bind to the goal sequence, however there can be no cleavage.
So the dCas9 protects the cleavage and modification website from energetic Cas9 on any given gene copy.
Notice that the quantity of lifeless Cas9 you add is crucial to the end result. So add a combination of Cas9 and dCas9 to your experiments. For instance, 1:1, 1:1.5, 1:2, 1:4, ratios, and many others.
Then, measure the dose response of your cells to dCas9, and plot the share proportion of homozygous, heterozygous, and wild-type clones towards the dose to choose the candy spot to your desired final result.
You too can monitor and quantify this optimization utilizing the BFP assay talked about above as a result of the dCas9 will enhance the proportion of wild-type clones, resulting in much less conversion of BFP to GFP.
High quality Management: Verify for Homozygous Clones
Deleterious on-target results that may happen throughout the modifying course of should be screened for.
For instance, one allele comprises the specified mutation, whereas the opposite is misplaced altogether.
To verify whether or not a homozygous clone is really homozygous, you are able to do long-range PCR, examine heterozygous single nucleotide polymorphisms (SNPs) on both facet of the goal sequence, and verify if they’re nonetheless heterozygous after you will have made your edit. [11]
Lack of SNP heterozygosity signifies that one allele has been deleted or translocated. Or there’s an undesirable insertion.
So beware!
The CRISPR iPSC Various to Immortalized Cells In Abstract
That’s rather a lot to summarize, however I’ll attempt!
Vital issues come up when utilizing immortalized cells, however CRISPR and cell reprogramming mix to create a substitute for immortalized cells.
We’ve explored the ability of this expertise and the best way to use it to check human illness and genetic variation.
And readers concerned in present genome modifying initiatives now have an arsenal of concepts, suggestions, assays, and sources, so as to add further layers of management.
Right here’s to extra impactful outcomes and discoveries from them.
Have you ever acquired something so as to add? Let me know within the feedback part under.
Need extra data? Watch the total presentation under.
References
- Medvedev SP, Shevchenko AI, and Zakian SM (2010) Induced pluripotent stem cells: issues and benefits when making use of them in regenerative medication. Acta Nat 2(2):18–28
- Rees HA and Liu DR (2018) Base modifying: precision chemistry on the genome and transcriptome of dwelling cells. Nat Rev Genet 19(12):770–88
- Liu LP and Zheng YW (2019) Predicting differentiation potential of human pluripotent stem cells: Prospects and challenges. World J Stem Cells 11(7):375–382
- Yu C, Liu Y, Ma T, et al. (2015) Small molecules improve CRISPR genome modifying in pluripotent stem cells. Cell Stem Cell 16(2):142–7
- Guo Q, Mintier G, Ma-Edmonds, M, et al. (2018) ‘Chilly shock’ will increase the frequency of homology directed restore gene modifying in induced pluripotent stem cells. Sci Rep 8:2080
- Ghanta KS, Chen Z, Aamir Mir, et al. (2021) 5′-Modifications enhance efficiency and efficacy of DNA donors for precision genome modifying. eLife 10:e72216
- Liu M, Rehman S, Tang X, Gu Okay, Fan Q, Chen D, and Ma W (2019) Methodologies for Bettering HDR Effectivity. Entrance Genet 9:691
- Skarnes WC, Pellegrino E, and McDonough JA (2019) Bettering homology-directed restore effectivity in human stem cells. Strategies 164–165:18–28
- Skarnes WC, Ning G, Giansiracusa S, et al. Controlling homology-directed restore outcomes in human stem cells with dCas9. bioRxiv
- Hodgkins A, Farne A, Perera S, Grego T, Parry-Smith DJ, Skarnes WC, and Iyer V (2015) WGE: a CRISPR database for genome engineering. Bioinformatics 31(18):3078–80
- Weisheit I, Kroeger JA, Malik R, Klimmt J, et al. (2020) Detection of Deleterious On-Goal Results after HDR-Mediated CRISPR Modifying. Cell Rep 31(8)
[ad_2]